How does a single-celled embryo know its top from its bottom? How can a single cell give rise to something with hair and a heart and only one head? This question of how an embryo sets up differences between cells during many divisions is an interesting and important one with wide ranging consequences.
In this study we are using the nematode worm C. elegans as our model organism since it uses both protein localization and asymmetric division to establish differences between cells as early as its first division.
The movie below shows the development of a C. elegans embryo from shortly after fertilization to first cleavage. Using this imaging technique, the only noticeable difference after first cleavage is the difference in size between the two daughter cells. What we don't see is that the distribution of proteins between the two cells is also very different, with the consequence that each daughter gives rise to a different set of differentiated cell types.
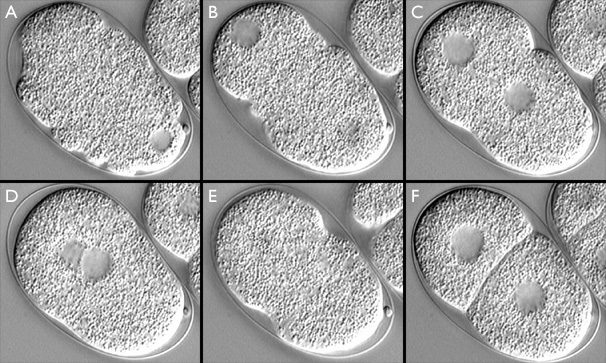
C. elegans embryo from just after fertilization to first cleavage. Anterior is to the lower right, posterior to the upper left. Clear areas indicate maternal and paternal pronuclei. (A) meiosis II, "ruffling" stage; (B) female pronucleus (lower right) begins to migrate toward male pronucleus (upper left); (C) "pseudocleavage"; (D) pronuclei meet and migrate to the center ("centration"); (E) first cleavage; (F) two-cell stage. This image has associated movies in two sizes. The movies will open in a new window sized as indicated. View the small version of the movie [500x620 pixels, 4.9 MB], or a larger version [700x820 pixels, 19.4 MB]. The image links to the small size movie.
The regulatory proteins of interest in this study, PAR and MEX proteins, are crucial for cell polarization in the C. elegans embryo. In particular, we are interested in the following proteins:
The PAR proteins are so named because in C. elegans, embryos mutant for these genes do not correctly partition cytoplasmic components and cleave symmetrically (partitioning defective). Embryos lacking MEX protein function differentiate an excess of muscle cells (muscle excess). PAR-3, PAR-6 and PKC-3 bind to form a complex that associates with the cortex of the cell. As they require each other in order to have an effect on the early embryo, they will be referred to collectively as the PAR-3 complex.
 |
 |
 |
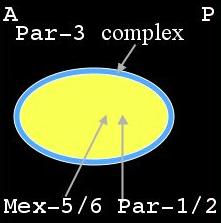
Distribution of PAR & MEX proteins immediately after fertilization during meiosis II |
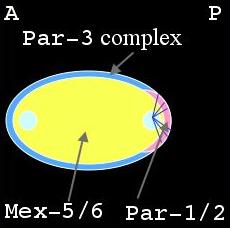
Distribution of PAR & MEX proteins after formation of sperm aster during establishment phase. |
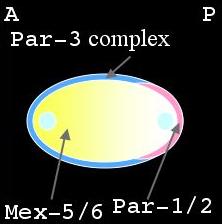
Distribution of PAR & MEX proteins during maintenance phase |
Shortly after fertilization during meiosis II, the PAR-3 complex is distributed on the cortex while PAR-1, PAR-2 and MEX-5/6 are uniformly distributed in the cytoplasm (left panel, above). As the male pronucleus forms a sperm aster, the PAR-3 cortical domain retracts, away from the male pronucleus, while PAR-1 and PAR-2 attach to the cortex in this region (middle panel). At the same time MEX-5/6 is being degraded in the posterior cytoplasm (right panel). The position of the male pronucleus determines the posterior end of the cell, while the female pronucleus is positioned at the anterior end. This is termed the establishment phase. Once this asymmetry is at its maximal value, it is maintained through to first division. This is the maintenance phase.
While the PAR amd MEX proteins are being redistributed in the early embryo, there is also movement of the cell cortex. The cell cortex includes an actin meshwork that myosin can bind to. Here we are referring to non-muscle myosin II which is the major protein that mediates actin-based contraction in non-muscle cells. From here on, we will refer to it simply as myosin.
In meiosis II, the embryo surface exhibits areas of high myosin density which indicate cortical contraction. These bright spots we call myosin foci, and they are revealed by a GFP-NMY2 fusion protein in the images below.
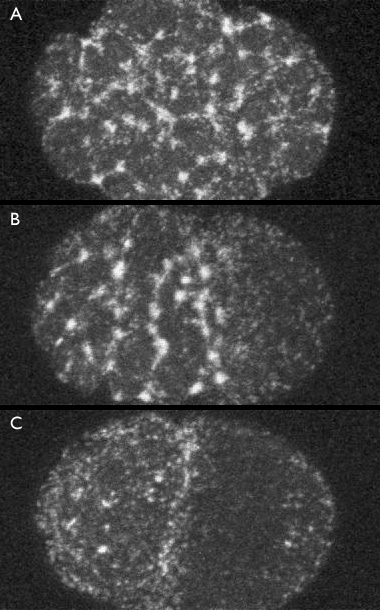
Distribution of myosin foci A) immediately after fertilization, B) during establishment phase, and C) during maintenance phase. Click the image to view the confocal time-lapse movie [550x480 pixels, 4.4 MB]. (courtesy of Ed Munro) from which these frames were selected.
In the figure above, Panel A shows that immediately after fertilization, during meiosis II, myosin foci are uniformly distributed around the egg surface. During the establishment phase, there is a clearing of the myosin foci from the posterior of the cell (Panel B). Once the embryo reaches the maintenance phase, the myosin mesh has cleared from approximately the posterior half of the cell (Panel C). Foci give way to a smooth mesh which itself quickly disappears as the cell enters mitosis. The flow of myosin [new window, 550x460 pixels, 4.4 MB]. toward the anterior of the cell is concurrent to the localization of Par and Mex proteins.
Recent work by Ed Munro has determined that the activity of the PAR and MEX proteins is coupled to these observed cortical dynamics and that cortical flow is essential for establishing these protein asymmetries.
There is strong evidence connecting cortical dynamics with the localization of proteins and that in turn, the proteins have an effect on cortical dynamics.
We reviewed the literature and past experimental work and compiled the information into a network diagram. This diagram indicates the interactions between the PAR & MEX proteins and the dynamics of cortical actomyosin. The purpose of this study is to explore the dynamics of this network in a two pronged approach. The first is to use computational techniques to explore interactions between these different players. The second uses various experimental techniques to clarify how the PAR & MEX proteins are regulating cortical actomyosin dynamics, and how actomyosin dynamics in turn affect the localization and activity of the PAR & MEX proteins.
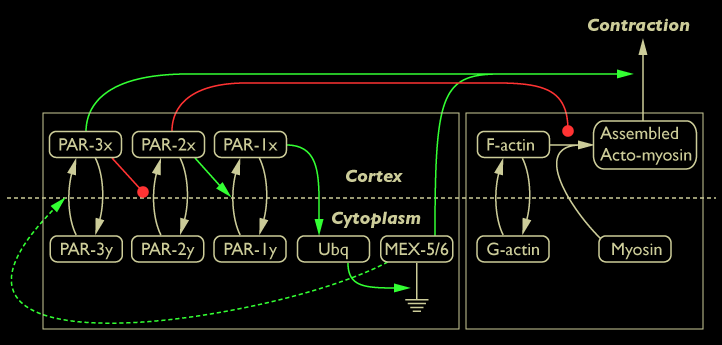
Diagram of interactions between PAR & MEX protein activity and cortical dynamics