developed by Ed Munro, Kristin Sherrard, Eliana Hechter and Adriana Dawes
Morphogenie is a flexible software program designed to explore how morphogenetic dynamics of single cells and multicellular tissues emerge from a local interplay between biochemical kinetics and cytoskletal mechanics. Click here to see an overview and description of how Morphogenie works. Click here to view a Java tutorial on simulating morphogenesis in a simplified version of adhesion. Or, click on the imags below to see some examples of simulations made using Morphogenie.
The pictures below link to QuickTime movies. The movies open in a new window sized as indicated.
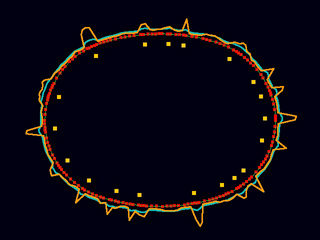
A model of the C. elegans cortex robustly reproduces the wild-type focal contractility cycle seen prior to polarization. (Ed Munro and Eliana Hechter)
[640 x 480 pixels, 12 MB]
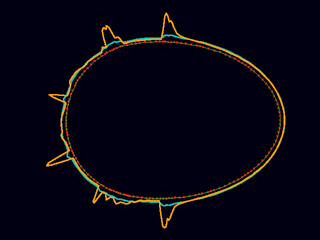
The same model responds to local weakening with a self-limiting global contraction to form a stable anterior cap. (Ed Munro and Eliana Hechter)
[640 x 480 pixels, 3.3 MB]
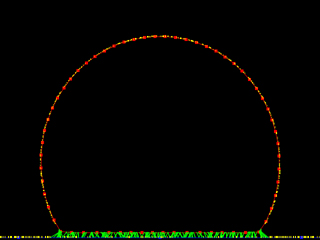
A dynamic "adhesion ratchet" causes a single cell expressing many adhesion receptors to spread on rigid external substrate coated with ligand. (Ed Munro)
[640 x 480 pixels, 911K]
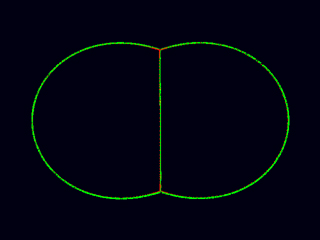
How dynamic adhesion and inhibition of myosin recruitment along domains of cell-cell contact could drive cell compaction and junction formation in embryonic blastomeres. (Ed Munro)
[589 x 806 pixels, 15.5 MB]
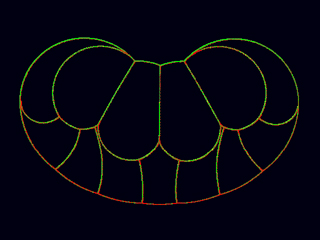
Simulation of endoderm invagination during gastrulation in the Ascidian embryo. (Kristin Sherrard and Ed Munro)
[640 x 480 pixels, 3.3 MB]
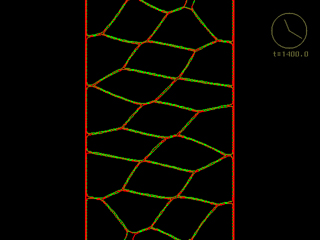
How localized protrusive, contractile and adhesive forces could drive convergent extension within a monolayer epithelial sheet. (Ed Munro)
[640 x 480 pixels, 4 MB]