
Research Professor
vicfoe@u.washington.edu
My research at the Center for Cell Dynamics focuses on two very different cell biological issues. For the last several years I have been working on i) understanding how the mitotic apparatus positions the contractile machinery that executes cytokinesis. And I have begun a new project—ii) investigating the effect DNA replication has on pol II transcription. My studies make use of marine invertebrate embryos (echinoderms or ascidians) and of Drosophila melanogaster. The Center for Cell Dynamics is located at Friday Harbor Laboratories, a marine field station and nature preserve on San Juan Island. I love doing modern cell biology in a rural setting, where wild animals are commonplace, and where the ocean teems with myriad strange life forms.
My favorite research instruments are my eyes. Modern probes and microscopes, human eyes, and the amazing interpretive abilities the human brain has evolved to make sense of the physical world together make up a powerful and versatile set of research tools. Though “merely descriptive” is today often used to disparage, most fundamental biological discoveries rest on careful description. Below I list some discoveries I’ve made looking through microscopes.
In my doctoral studies (begun at UT Austin in the laboratory of Hugh Forrest and completed at UW in the laboratory of Charles Laird) I used transmission electron microscope (TEM) imaging of chromatin preps from milkweed bug (Oncopeltus fasciatus) embryos to study transcriptional activation. I showed that analysis of TEM data can provide information on relative levels of transcriptional activity that agree well with biochemical data obtained by RNA-polymerase runoff assays. Analysis of the organization (length and distribution) of nascent transcripts along the chromatin revealed discrete sites of transcriptional initiation and termination and was the first study to show that the average unit of non-ribosomal transcription was well over an order of magnitude longer than would be needed to encode a single protein (1). Analysis of the organization and length of nascent transcripts revealed that in some transcription units transcripts were cleaved at specific sites prior to their release from the underlying DNA, thus providing the first glimpse of introns and their excision from hnRNA (2,3). I discovered that while DNA transcribed by RNA polymerase II remains organized in nucleosomes, ribosomal chromatin transcribed by RNA polymerase I has a different and non-nucleosomal structure, even prior to transcription (1,4).
In subsequent studies, using the same EM imaging technique used for visualizing interphase chromatin, I discovered that the condensed holokinetic chromosomes of the milkweed bug are organized into transcriptionally-inert looped domains of highly condensed chromatin, with the loops densely arrayed along a negatively-staining central axis (3); images also shown in (5).
As a post doc, in the laboratory of Bruce Alberts at UCSF, I characterized the pre-gastrula morphogenetic and cytoplasmic movements of Drosophila embryos and generated a high-precision timetable of early Drosophila embryogenesis [opens new window, 760 x 500 pixels, 156 KB]. I showed that in this syncytial embryo the cytoskeletal reorganizations that drive the early morphogenetic movements (nuclear migration, formation and breakdown of pseudo-cleavage furrows and cellularization) are all precisely synchronized with the mitotic cycle (6,7).
Using epi-fluorescence microscopy I studied the changes in morphology that embryonic Drosophila nuclei undergo during transcriptional activation and cell differentiation. In the course of this work I discovered that depriving Drosophila embryos of oxygen, or inhibiting oxidative phosphorylation by other means, rapidly plunged the embryos into a reversible state of suspended animation, which they can survive for several days (8). During anoxia I found that the interphase chromosomes had condensed onto the inner surfaces of intact but swollen nuclear envelopes. Each condensed chromosome looks like a thin convoluted snake with its telomeric tails pinned at the basal pole of the nucleus, facing the embryo interior, and its kinetochore pinned at the nuclear apical pole (image shown immediately below illustrates this). This finding implies that the interphase nucleus is organized such that chromosomes retain their telophase orientation, even while chromosome loops decondense for transcription or replication, and even when the nucleus as a whole rotates 90° between telophase and interphase (8). Suspended animation brought about by inhibiting oxidative phosphorylation is now known to be a highly conserved protective mechanism, physiologically similar to the hypometabolism of hibernation, diapause and estivation, and may ultimately find use as a medical intervention strategy during surgery, ischemia and for organ preservation (9).
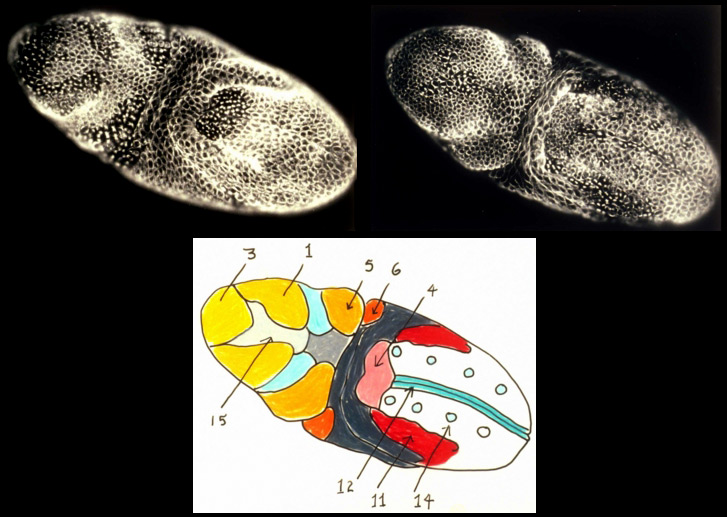
In the syncytial Drosophila embryo, mitoses proceeds with global synchrony during the first 13 nuclear cycles. However following cellularization during mitotic cycle 14, I discovered that, examined simply from the vantage of cell cycle length, the post-gastrula Drosophila embryo is partitioned into a fate map of 25 distinct, bilaterally symmetric, "mitotic domains". In addition to a characteristic cell cycle length, cells in different mitotic domains often exhibit cell properties distinct from those of cells in adjacent mitotic domains, properties such as orientation of division, a characteristic cell shape, and specific type of motility. The mitotic domains of cycle 14 are in many cases further subdivided into smaller mitotic domains during mitoses 15 and 16. My studies provided a detailed mitotic domain atlas [new window, 840 x 540 pixels, 104 K] and a cell division timetable [new window, 800 x 540 pixels, 96 K] for the Drosophila embryo's 14th nuclear division. The same tight coupling between cell cycle and cell determination is observed, for example, in ascidian embryos, but the means by which this coupling occurs remains to be discovered.
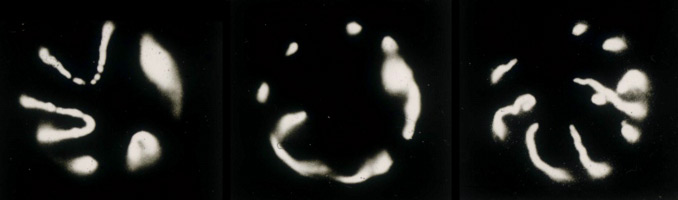
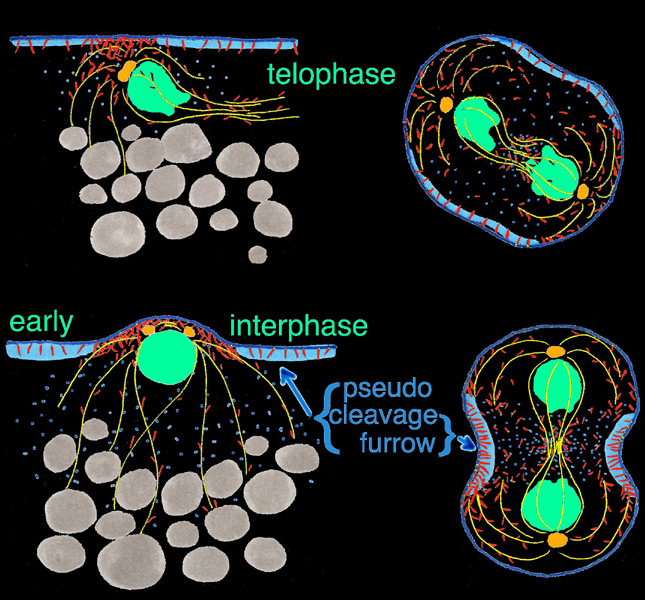
Using scanning laser confocal microscopy (LSCM) Christine Field and I studied the temporal and spatial reorganizations of microtubules, F-actin and cytoplasmic myosin II in syncytial blastoderm-stage Drosophila embryos (12). This paper's Figure 2 [opens new window, 800 x 500 pixels, 428 KB] illustrates in surface view the rapid cyclic reorganizations of cortical F-actin and myosin II that occur during syncytial cycles 9 -11, and show that microtubules somehow work against co-localization of F-actin and myosin II in the syncytial cortex. The timelapse video of GFP myosin regulatory light chain [opens new window, 560 x 400 pixels, 59.5 MB] shows in live Drosophila embryos what Figure 2 and the cross-sectional image immediately below show for fixed embryos — regulation of cortical myosin II by the mitotic cycle and by proximity to microtubule arrays. Stax, a segmentation and 3-D projection software tool created by Center for Cell Dynamics scientific director Garrett Odell, allows the rendering of hundreds of serial confocal micrographs into deep three-dimensional images. These tools allow us to visualize the 3-D distribution of microtubules, centrosomes, nuclei, F-actin, myosin II and yolk particles in syncytial Drosophila embryos as movies and interactive virtual reality images (click here to see Drosophila 3-D reconstruction samples).
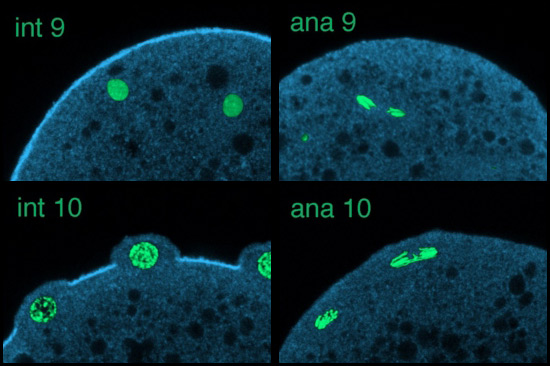
Drug studies and the rapid cytoskeletal reorganizations we observed imply interactions between microtubules and F-actin, between microtubules and myosin II, and between each of these and the mitotic oscillator. Specifically our data suggested i) that F-actin binds and is transported along microtubules towards microtubule plus ends; ii) that microtubule arrays or something closely associated with them causes localized loss of myosin II from the adjacent cortex; and iii) that cell cycle phase determines the levels of both F-actin and myosin II that associate with the cortex, with both proteins accumulating on the cortex from telophase through interphase, but beginning to leave the cortex during metaphase and reaching a nadir at anaphase. Our findings for syncytial-stage Drosophila embryos suggested how contractile ring assembly might be temporally and spatially regulated during standard cytokinesis (i.e. in cells where cell division follows each nuclear division, which does not occur in the syncytial-stage fly embryo).
I moved to Friday Harbor in 2002 to work on cytokinesis with George von Dassow. Clever micromanipulation experiments, most preformed two decades ago by Rappaport and Hiramoto, have established the “rules” by which the mitotic apparatus determines the position of the cleavage furrow, but this work preceded confocal microscopy and modern probes. As had the classical work before us, we chose echinoderm embryos for our studies, but we have undertaken to perfect fixation and staining protocols and to develop live probes in order to study the cytoskeletal dynamics and signaling events that underlie the rules of cytokinetic furrow specification. My discovery of a unique population of nocodazole-resistant astral microtubules which form during anaphase, the accumulation at their plus ends of phosphorylated myosin, and computer simulations by Garrett Odell comparing MKLP1 movement on stable vs. dynamic microtubules combine to suggest a new model for furrow stimulation. These and our related discoveries are summarized under Research Report for Cytokinesis.
|
|
|
|
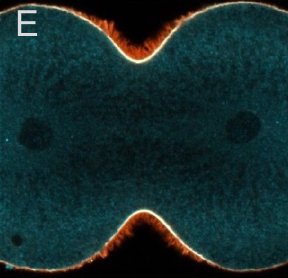
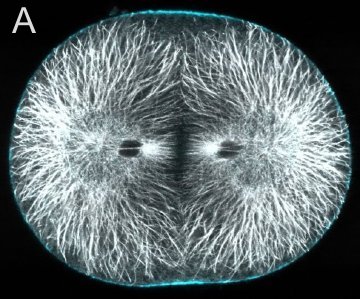
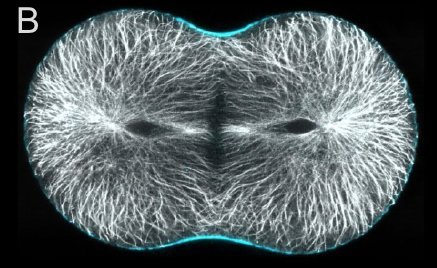
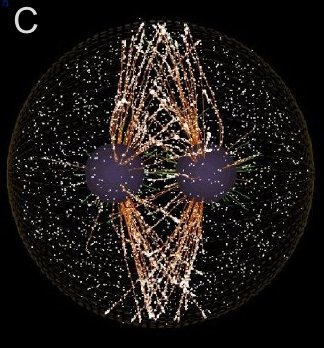
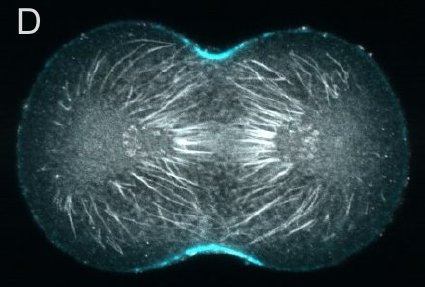
A and B are confocal micrographs of a green urchin zygote, showing microtubules in white, and in blue myosin regulatory light chain phosphorylated on serine19 (an activating phosphorylation). D shows in a purple urchin zygote the subset of microtubules that resist depolymerization (white), and serine19 phosphorylated myosin RLC (blue) accumulating where these stable microtubules contact the cortex. E shows in a cleaving sand dollar zygote, myosin heavy chain (dark blue) and F-actin (orange); the region where both proteins concentrate is white.