Biology department at Universidad de Antioquia
hoyose@u.washington.edu
CCD Research Intern
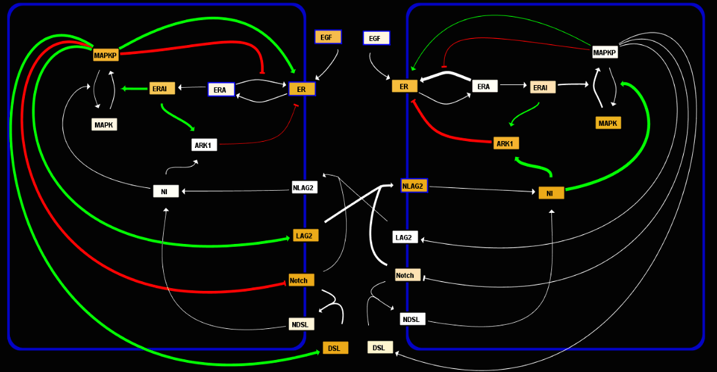
I became interested in the Center for Cell Dynamics while I was a Biology undergraduate student seeking to learn about developmental gene networks; looking at their webpage, I realized that they periodically teach apprenticeships where they give students from all over the world the opportunity to learn not only experimental, but also advanced computational modeling techniques to study how developmental networks are capable to establish complex patterns of molecular concentrations throughout the embryo. I was fortunate when I was accepted to the 2006 Research Apprenticeship on Computational Modeling of Biological Networks at the CCD, where I worked, under the advice of Dr. Kerry Kim and in collaboration with Marie-Anne Felix and Josselin Milloz from the Jacques Monod Institute, on a project intended to reconstitute the known molecular interactions between the canonical EGFR/Ras/MAPK and Notch pathways during vulva formation in Caenorhabditis elegans. This network is responsible to determine a conserved 3o3o2o1o2o3o pattern of differentiation among the six vulval precursor cells (VPCs), which after following four rounds of mitotic divisions give raise to the morphology of the vulva.
After the apprenticeship, the CCD gave me the opportunity to continue working on this project for a longer period of time, which eventually became my undergraduate thesis. Our purpose with this study is to explore in detail the dynamics of this system that allows it to robustly determine the pattern of cellular fates conserved among different Caenorhabditis species; besides this, our model let us mechanistically distinguish between different modes of induction, which affect the system’s tolerance to developmental noise and environmental perturbations. Moreover, using this model we can explore how the network acts as a 3-way switch to stabilize an isolated VPC in one of the three possible cellular fates under different concentrations of the inductive signal. Finally, by reproducing different mutations done in the C. elegans N2 wild isolate, and in several distinct Caenorhabditis species, we plan to unmask quantitative variations of this network accumulated throughout evolution; by finding the relative position of different species in parameter space, we plan to calculate evolutionary distances that can be further compared with current phylogenies established by genome sequence comparisons.
In the future, I would like to study how developmental networks allow the evolution of the nervous system, and the interplay between molecular and neural networks in the context of high order brain functions such as learning and memory.
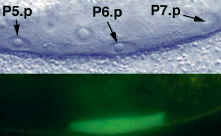
egl17::GFP Ambros (1999)
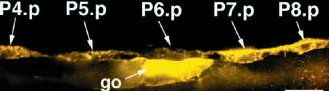
lip1::GFP Ambros (1999)