The first time I saw a time-lapse movie of a cell dividing I was simply amazed. It was sublimely beautiful. The more I've learned about cytokinesis and other dynamic cellular behaviors, the more puzzled and fascinated I've become. These behaviors are the consequence of millions of local interactions among molecules that have no knowledge of what they are doing in the context of the whole. No one is driving the bus! This is the greatest mystery I have ever encountered. For the past four years, I have been working at the Center for Cell Dynamics (CCD) under the guidance of Dr. Garret Odell and Dr. Ed Munro to learn different computational approaches to understanding how complex cellular behaviors emerge from local interactions among biochemical regulators and cytoskeletal proteins.
I have been particularly drawn to agent-based models of cytoskeletal dynamics. My current work involves the construction of a 3D agent-based computer simulation of the actin rich cortex on the order of a few microns in diameter. The goal is to use this model to explore how meso-scale contractile and viscoelastic properties of a patch of cortex emerge from the micro-scale dynamical interactions of actin-filaments, cross-linkers, regulating proteins, and myosin motors. At every timestep, each cytoskeletal part in the simulation figures out what to do next by numerically solving its zero-inertia Newtonian force/torque balance equations of motion. We are calibrating our model against the measured viscoelasticity of networks assembled from purified components in vitro. I am now running simulations simultaneously on the ~450 processors of the CCD's linux cluster to explore how the viscoelastic properties of this complex filament network depend on densities and properties of different cross-linkers and on filament assembly dynamics.
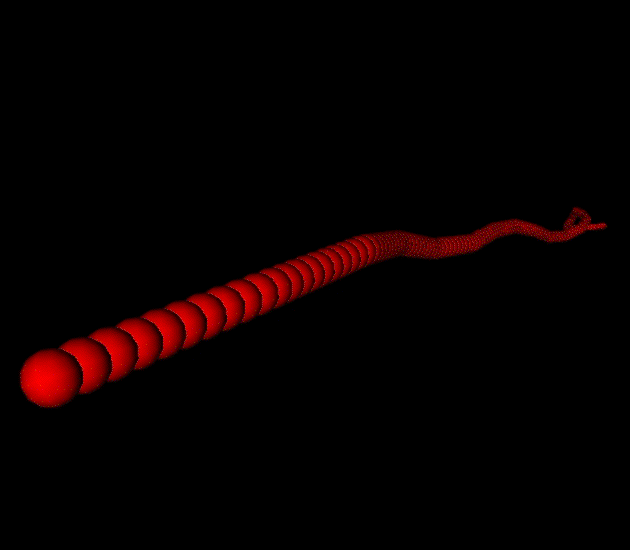
We have biophysically realistic simulated actin filaments which have been tuned to match experimentally determined values for its persistence length, and theoretically derived elastic modulus, and relaxation times when undergoing deformation. We also have devised a generic framework for cross-linker proteins, which allow us to specify their length, relative binding affinity, force dependent breakage, and preferred angles for binding to the filament. In the movies shown the cross linker is alpha-actinin.
 |
 |
 |
|---|---|---|
A single filament undergoing thermal writhing. |
Forces and Torques acting on two filaments undergoing thermal motion and bound together by linkers. |
Steric Hinderance of filaments |
We are now calibrating the networks behavior to match one-point and two-point microrheology experiments in vitro.
 |
 |
 |
|---|---|---|
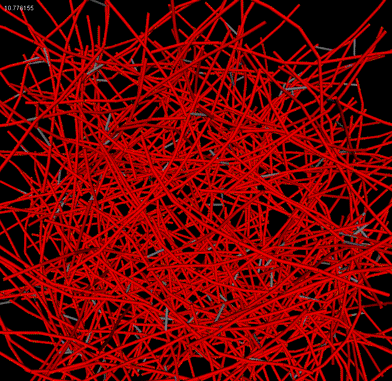
| Patch of cortex |
Downard flythrough of cortex |
Microrheology of filament network |
 |
||
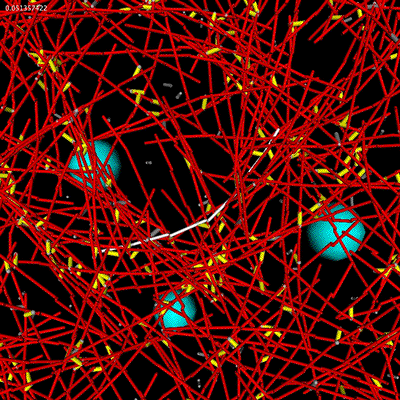
| Microrheology of filament and linker network. |
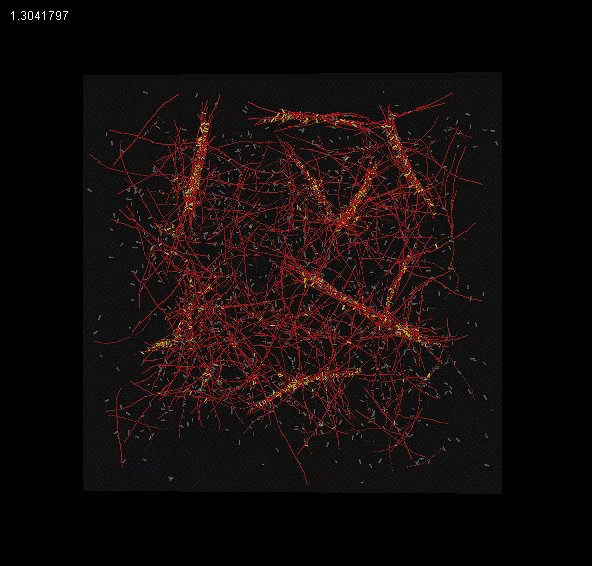
The meshwork currently exhibits the qualitative features of bundling for certain types of linkers and gel effects at high linker concentrations.
 |
 |
|
|---|---|---|
| Filament Bundling |
Gel effects at high linker concentration |
Bead tracking |
I am also interested in oligomeric protein networks and how their transient structures arise to form diffusion traps and scaffolds in the cell. I wrote a generalized agent-based simulation that makes it easy to specify unique properties (diffusivity, number of binding sites, etc) for individual monomers and the properties of the bonds between them. We can endow these proteins with a specific geometry to approximate the orientation of their binding sites or represent them in their simplest form as spheres. Dr. Munro and I are currently using this model to investigate the properties of the Par protein oligomer network that forms near the cortex in polarizing C. elegans embryos.
I have come to believe this style of modeling can actually compute rigorously how the cytoplasmic streaming and cell cleavage behavior of a cell's cortex springs from the self-assembling action of its molecular parts. The future plans for this model are to use it to understand other biological phenomena including mechanisms of signal transduction through cytoskeletal elements, initiation and organization of filopodia and other protrusive structures, phagocytosis and other aspects of cellular motility.
For some earlier projects I developed an interactive diffuser model and a microtubule polymerization cell automata.
 |
 |
 |
|---|---|---|
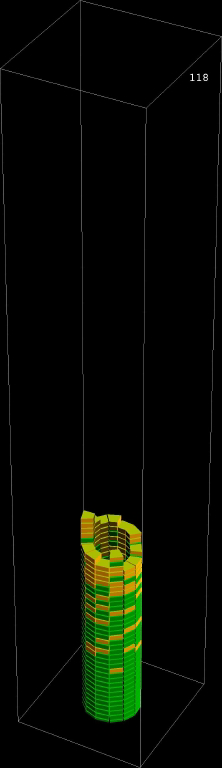
| Microtubule polymerization cell automata |
Microtubule polymerization cell automata with a tip tracking protein |
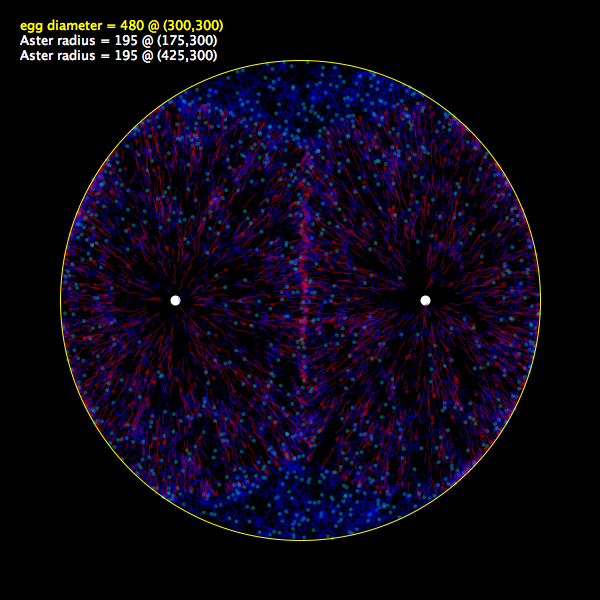
Interactive diffusion model |
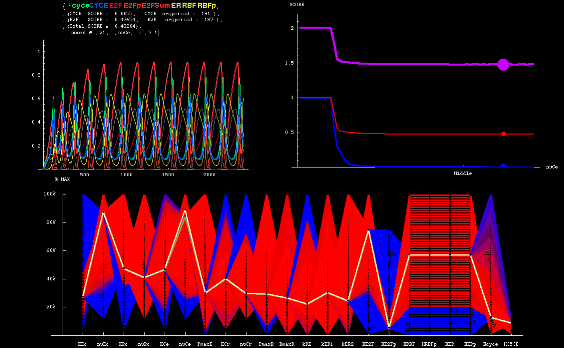
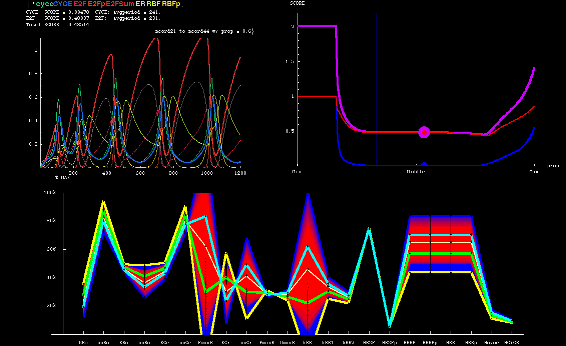
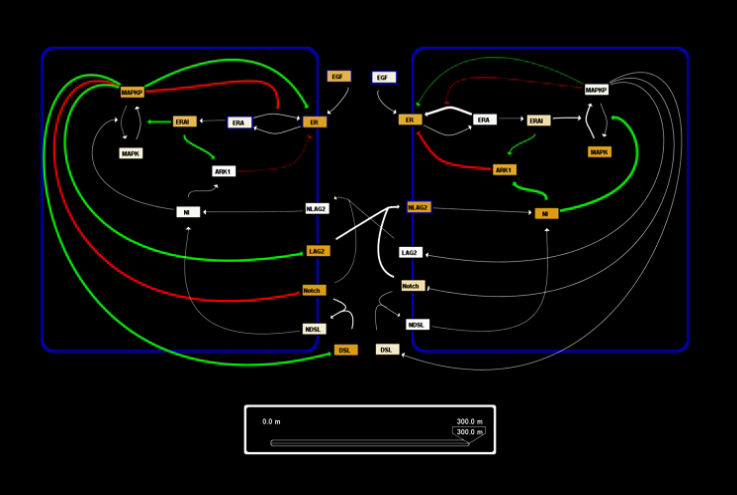
As part of a collaboration between Dr. Bruce Edgar of the Fred Hutchinson Cancer Research Center, and Dr. Kerry Kim and myself at the CCD, we modeled the endocycle control circuit in Drosophila. The endocycle is a modified mitotic cycle in which cells replicate, and thus increase their DNA, but do not divide. Cells thereby become larger and achieve a higher metabolic output as the higher ploidy leads to increased rates of transcription and higher protein levels. We explored the endocycle's intrinsic tuner involving kinetic parameters that control its frequency. I developed Mathematica software to analyze and visualize the high dimensional parameter space of the network.
 |
 |
 |
|---|---|---|
| Visualization of Robustness |
Visualization of path in high dimensional parameter space between two good parameter sets |
Network dynamics viewer |
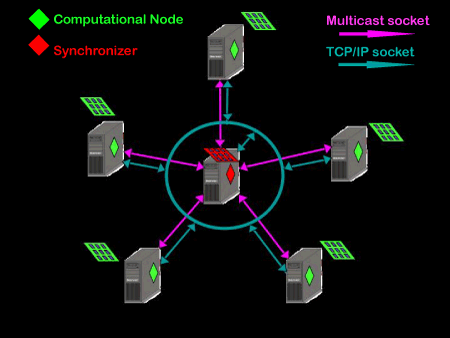
As many of the simulations at the CCD require enormous computational resources it is desirable that they be able to be run in parallel. In practice, writing a simulation to run in parallel can be quite challenging, as some tasks do not lend themselves easily to it. Assuming it can be partitioned properly the simulation will go faster when it is distributed amongst many computers, however, the speed at which an individual computer can get the next data segment it needs can be such a bottleneck to the overall performance that the advantages of going parallel are lost. My first project involved designing and writing a slick mechanism for distributing the data to exploit the benefits of parallel processing, by minimizing the hindering effects of the data bottleneck. The heart of this mechanism is a multicast network that cleverly polices the traffic between computers. One computer acts to synchronize the behavior of all others, via polling and handling requests loaded in a queue. Individual computers act as computational nodes on the network, listening for data they need, modifying it appropriately, and then multicasting it back onto the network, when permission is given by the synchronizing computer. The end result is the data "hovers" around the network, without needing to be gathered and resent redundantly between the computational nodes. This scheme increases the efficiency of the data flow, reduces the collisions between packets, and has scalability.
 |
|---|
| Multicast network schematic |