Duane Compton
The spindle is a complex microtubule-based structure responsible for chromosome movement and segregation (Figure 1). Minus ends of spindle microtubules are focused at spindle poles which are the sites of chromatid convergence upon anaphase onset. In many cell types centrosomes are located at spindle poles as a consequence of their role in microtubule nucleation. However, centrosomes are not necessary for spindle pole organization or function, as demonstrated by the lack of centrosomes in some cell types and experiments where centrosomes have been eliminated from cells. Instead, microtubule minus-ends are focused at spindle poles by non-centrosomal proteins, including the structural protein NuMA and microtubule motor proteins dynein (with its activating complex dynactin), HSET and Eg5. These proteins work in concert to focus microtubule minus-ends and tether centrosomes to the spindle pole through microtubule crosslinking and/or motor activity.
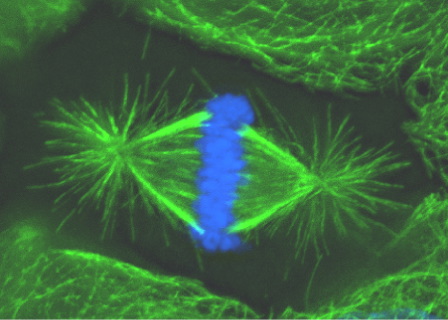
Figure 1. During metaphase of mitosis chromosomes (blue) align at the spindle equator. Minus ends of microtubules (green) within the spindle are focused at two spindle poles.
To investigate mechanisms of microtubule focusing at spindle poles we utilize a combined approach that involves in vitro biochemistry, in vivo cell biology, and computer modeling. In a cell free system microtubules are organized into asters (Figure 2) by the structural protein NuMA and motor proteins dynein (with its activating complex dynactin), HSET and Eg5. Extensive biochemical characterization of these microtubule asters indicates that the five non-centrosomal proteins mentioned above are the only proteins responsible for microtubule aster organization (Mack et al., 2001). Experiments where each protein has been inhibited alone or in various combinations has revealed important relationships between these proteins. For example, the minus end-directed motor activity of either dynein or HSET is sufficient for microtubule organization. Also, there is antagonism between motors of opposite polarity (Mountain et al., 1999).
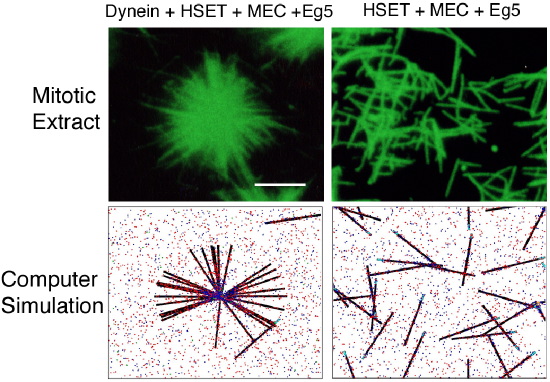
Figure 2. Microtubules in a mitotic extract form asters in the presence of dynein, HSET, NuMA (MEC), and Eg5 activities, but are randomly arranged if dynein is depleted. Computer simulation of these proteins yields similar results to the mitotic extract.
To understand how each protein contributes to microtubule organization and the nature of the relationships among these proteins we have utilized computer simulation (Figure 2). Computer simulation allows very fine-grained control of the microtubule motor and crosslinking activities. That work has led to a mechanistic model that states that the combination of two aggregate properties, Net Minus end-directed Force and microtubule Crosslinking Orientation Bias, determine microtubule organization. This model utilizes motor and non-motor proteins, accounts for motor antagonism, and predicts that alterations in crosslinking orientation bias should compensate for deficits in motor activity. We tested that prediction in the mammalian mitotic extract and, consistent with the model, found that increasing the contribution of microtubule crosslinking by NuMA compensated for the loss of Eg5 motor activity. Thus, this model proposes a precise mechanism of action of each non-centrosomal protein during microtubule aster organization, and suggests that microtubule organization in spindles involves both motile forces from motors and static forces from non-motor crosslinking proteins.
cytomechanical modules 2003 • back