Andrea Brand
A simple way to generate two different cell types upon division is through the asymmetric segregation of cell fate determinants, a mechanism that is used in organisms as widely divergent as bacteria and mammals. Asymmetric segregation is used in yeast to distinguish mother cells from their daughters, and in C. elegans, to differentiate the sister cells arising from the first embryonic division. Asymmetric cell division is also observed in the developing nervous system of both vertebrates and invertebrates. In the Drosophila nervous system, a number of cell fate determinants segregate to the GMC upon division. One is the homeodomain transcription factor, Prospero, named after the wizard who controls the fate of the other characters in Shakespeare's "The Tempest". Prospero activates GMC-specific genes such as even-skipped and ftz and represses neuroblast-specific genes such as asense and deadpan.
The asymmetric localisation of determinants is an elegant mechanism for rapidly switching from a stage of proliferation, when similar daughter cells are born (Figure 1, left), to a stage of diversification, when different daughter cells are generated (Figure 1, right). For example, in epithelial cells Prospero localises basolaterally but is segregated equally to both daughter cells at division. Neuroblasts also localise Prospero basally but, as they divide perpendicular to the epithelial layer, only one of the daughter cells inherits Prospero. Coordination of the mitotic spindle with the apico-basal axis of the cell appears to be a prerequisite for successful asymmetric segregation. We have shown that during pro/metaphase the neuroblast mitotic spindle rotates 90°, so as to align itself along the apico-basal axis of the cell and asymmetrically segregate Prospero.
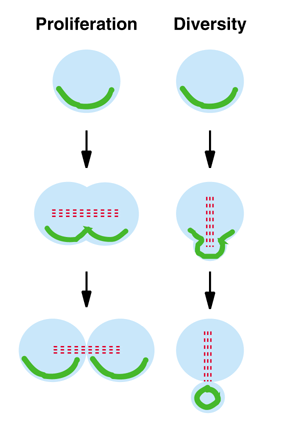
Figure 1
Prospero protein is localized to the basal cortex of neuroblasts by the adapter protein Miranda, a novel coiled-coil protein (named after Prospero's daughter in The Tempest). Prospero mRNA is also basally localised. Miranda binds directly to Prospero protein and also, we have shown, to Staufen, a double-stranded RNA binding protein, which in turn binds prospero mRNA. When neuroblasts divide, only the basal daughter cell (the GMC) inherits Miranda and its cargo of Prospero protein and prospero mRNA. Miranda then releases Prospero protein, which translocates to the GMC nucleus.
F-actin appears to play a key role in the asymmetric localisation of cell fate determinants in yeast and worms. Similarly, in Drosophila when F-actin is depolymerised by treatment with latrunculin, Prospero is released from the cortex. The mechanism by which Prospero (and its adapter Miranda) localise to the basal half of the cell cortex is unclear, as F-actin is uniformly distributed around the cortex.
The dynamics of asymmetric localisation and the dependence on F-actin suggest a role for myosin motor proteins in asymmetric cell division. We have found that cytoplasmic Myosin II, or Zipper, plays an integral role in localising determinants in neuroblasts. Myosin II is the homologue of C. elegans NMY-2, which is required to localise PAR proteins asymmetrically at the cortex of the one-cell embryo, revealing a conserved role for myosin II motors in mediating actin-dependent asymmetric segregation. Myosin-II has also been shown also to localise b-actin mRNA to the leading edge of fibroblasts.
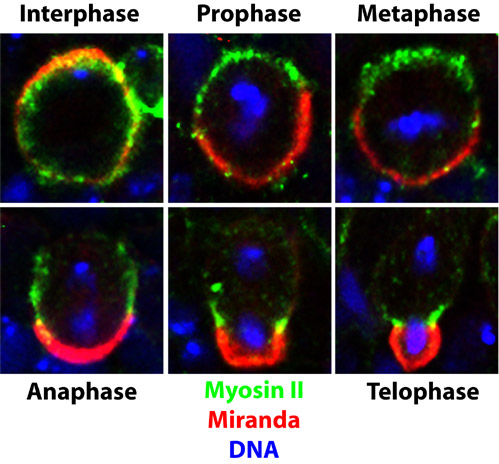
Figure 2
In contrast with NMY-2, which is localised uniformly around the cortex of the worm zygote, we find that Drosophila Myosin-II is asymmetrically localised in neuroblasts (Figure 2). Interestingly, Miranda and Myosin do not overlap extensively in neuroblasts. As Myosin moves to the cleavage furrow during anaphase and telophase, it appears to "push" Miranda into the GMC. I would like to discuss the possible mechanisms by which Myosin-II is localised, and the means by which Myosin might localise Miranda. One clue is that Drosophila Myosin-II interacts directly with the tumour suppressor, Lethal giant larvae. In yeast, the Lgl counterparts Sro7p and Sro77p exhibit strong genetic interactions with the myosins Myo1 and Myo2. Lgl is itself required for asymmetric cell division in neuroblasts and in yeast has a role in exocytosis.
cytomechanical modules 2003 • back